
Simulation of signal pathways and gene-regulatory networks often requires a stochastic formulation of the reaction kinetics in order to correctly capture the influence of small numbers of molecules and relevant fluctuations. However, with increasing number of molecules, the computational effort drastically increases.
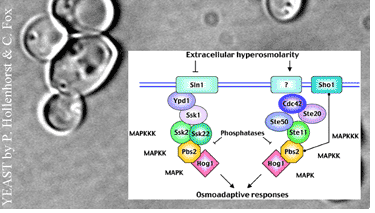
In contrast, metabolic networks with large numbers of molecules and continuously occurring reactions are successfully modelled by the deterministic formulation of chemical reaction kinetics (based on the law of mass action). The aim of the project is to develop a mathematical theory and adaptive numerical schemes for the analysis and simulation of coupled stochastic and deterministic models of biochemical reaction systems. Moreover, we are interested in efficient algorithms for directly solving the chemical master equation.
Within this project, we cooperate with experimentally working groups at the Freie Universität Berlin (Prof. Regine Hengge![]() , Microbiology) and the University of California, San Diego (Prof. Alexander Hoffmann
, Microbiology) and the University of California, San Diego (Prof. Alexander Hoffmann![]() ), as well as with theoretical oriented groups at the Zuse Institute Berlin (Prof. Peter Deuflhard
), as well as with theoretical oriented groups at the Zuse Institute Berlin (Prof. Peter Deuflhard![]() ) and the Freie Universität Berlin (Prof. Christof Schütte
) and the Freie Universität Berlin (Prof. Christof Schütte![]() ).
).